The pmemd.cuda GPU Implementation
Por um escritor misterioso
Descrição

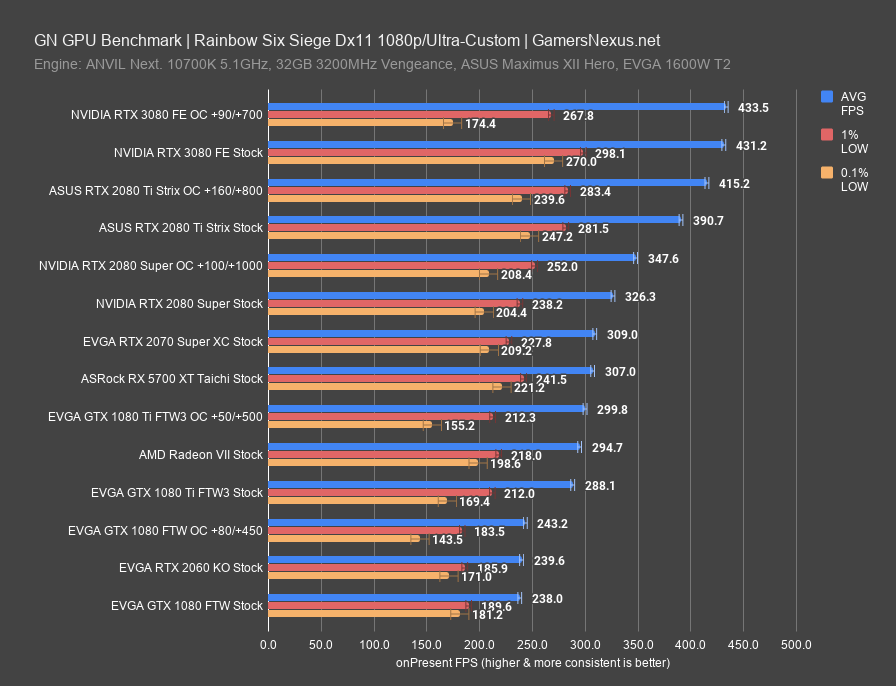
Performance of Amber 18, NAMD version 2.13 and GROMACS 2018 measured as

Nvidia Cuda Apps Jun27 11

Full article: Solvated and generalised Born calculations differences using GPU CUDA and multi-CPU simulations of an antifreeze protein with AMBER
The pmemd.cuda GPU Implementation

GUI for setup and control of Amber18/20 molecular dynamics simulations

Statistical machine learning for comparative protein dynamics with the DROIDS/maxDemon software pipeline - ScienceDirect

Toward Fast and Accurate Binding Affinity Prediction with pmemdGTI: An Efficient Implementation of GPU-Accelerated Thermodynamic Integration.

Schematic representation of the main components of the MIST library.
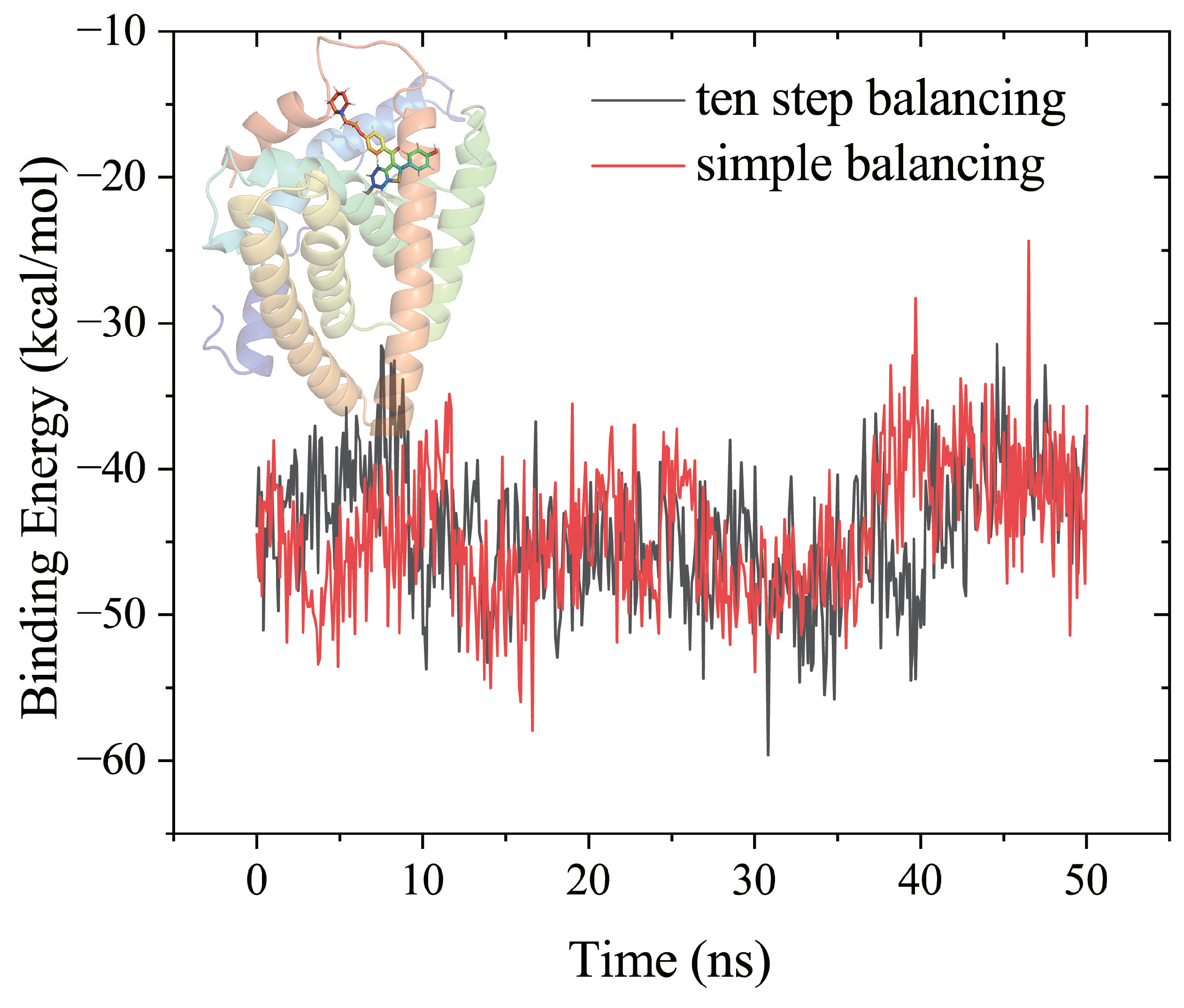
Biomolecules, Free Full-Text
AMBER14 & GPUs

Fast Implementation of the Nudged Elastic Band Method in AMBER

Full article: Solvated and generalised Born calculations differences using GPU CUDA and multi-CPU simulations of an antifreeze protein with AMBER

GPU-Accelerated Implementation of Continuous Constant pH Molecular Dynamics in Amber: pKa Predictions with Single-pH Simulations
de
por adulto (o preço varia de acordo com o tamanho do grupo)